Sequencing Immunoglobulin Genes to Detect Residual Traces of Cancer
Overall, this new method is a promising approach for the detection of minimal residual disease in B cell acute lymphoblastic leukemia (B-ALL) patients, particularly given its strong performance and ease of use compared to current methods.
B cell acute lymphoblastic leukemia (B-ALL) is a cancer that develops in white blood cells called lymphocytes. Following treatment, one important indicator for prognosis is whether any remaining leukemia cells, termed minimum residual disease (MRD), can be detected. This information can be useful for reinitiating or intensifying treatment, but current test methods have limitations that hamper their broad implementation. In a recent report, Dr. Harlan Robins in the Public Health Sciences Division demonstrates an accurate and sensitive new method for detecting minimal residual disease via high-throughput sequencing, which could significantly improve clinical prognostication in B-ALL patients.
Current methods for detecting MRD are flow cytometry and quantitative PCR, which are able to detect MRD down to a level of roughly 1 in 100,000 cells. However, "monitoring leukemia and lymphoma after treatment has been tricky," said senior author Dr. Robins. "The methods are disease dependent, hard to standardize, and have limited sensitivity. We have a new molecular test that addresses and solves each of these issues, and our paper applies this new method to B-ALL." This new molecular test utilizes high-throughput sequencing of complementary-determining region 3 (CDR3) of the immunoglobulin heavy chain gene, IGH. Unique rearrangements in B cells of the V, D, and J segments of this gene produces unique antibodies, but clonal expansion of leukemic B cells results in overrepresentation of a specific rearrangement in the blood. By measuring rearrangements of the V, D, and J segments through sequencing, clonal populations representing MRD can be detected.
To evaluate the utility of this assay, the authors compared their IGH sequencing approach with the standard flow cytometry method performed at the University of Washington. This was done using pre- and post-treatment paired samples from 98 B-ALL cases from the Children’s Oncology Group AALL0932 trial. Said Dr. Robins, "This allowed a direct comparison of our approach to the state of the art measurement of residual disease from one of the premier monitoring labs in the country." In 92 of the 98 pre-treatment samples, the authors were able to identify the unique, recombined IGH gene sequences that represented each patient’s clonal, neoplastic B lymphoblasts. The presence of these original clonal rearrangements could then be looked for in post-treatment samples to identify MRD.
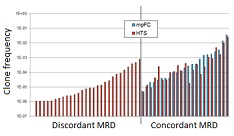
Comparing the two methods, both detected MRD in 23 samples and did not detect MRD in 40 samples. However, there were 28 cases where MRD was detected by the IGH sequencing method but not flow cytometry (see figure). There were no cases where flow cytometry detected MRD but sequencing did not. Though a few of these detection-discordant samples were within the detection limit for flow cytometry, the authors estimated that their method had a 10-fold lower detection limit for MRD, at around 1 cell in a million. Since early detection of MRD is important for personalized treatment of B-ALL, this more sensitive detection could potentially improve patient prognostication. Said Robins, "These results are very encouraging, and suggest that our ClonoSeq technology can be translated effectively to the clinic."
Overall, this new method is a promising approach for the detection of minimal residual disease in B-ALL patients, particularly given its strong performance and ease of use compared to current methods. Additional studies will be needed, however, to understand the clinical implications of this more sensitive test. Said Robins, "Moving forward, we need to extend the study to much larger cohorts to evaluate differences in clinical outcome for low level residual disease (levels not detectable by presently employed methods). Additionally, we are in the process of applying the technology to many other types of leukemia and lymphoma. We hope to see a rapid uptake of ClonoSeq in the diagnostic market over the next couple years."
Illustration: Comparison of the clone frequencies identified by high-throughput sequencing (HTS, red) versus multi-parametric flow cytometry (mpFC, blue), for detection of minimal residual disease (MRD) in 29-day post-treatment B cell acute lymphoblastic leukemia cases (x-axis). The two methods both detected MRD for 23 samples (right), while for 28 samples MRD was detected using HTS but not mpFC (left). –Dr. Harlan Robins.
Read more…
Fred Hutchinson Cancer Research Center News Release (08/18/14)
Abstract (Clinical Cancer Research; 20(17), 4540-4548 (09/01/14))
Current methods for detecting MRD are flow cytometry and quantitative PCR, which are able to detect MRD down to a level of roughly 1 in 100,000 cells. However, "monitoring leukemia and lymphoma after treatment has been tricky," said senior author Dr. Robins. "The methods are disease dependent, hard to standardize, and have limited sensitivity. We have a new molecular test that addresses and solves each of these issues, and our paper applies this new method to B-ALL." This new molecular test utilizes high-throughput sequencing of complementary-determining region 3 (CDR3) of the immunoglobulin heavy chain gene, IGH. Unique rearrangements in B cells of the V, D, and J segments of this gene produces unique antibodies, but clonal expansion of leukemic B cells results in overrepresentation of a specific rearrangement in the blood. By measuring rearrangements of the V, D, and J segments through sequencing, clonal populations representing MRD can be detected.
To evaluate the utility of this assay, the authors compared their IGH sequencing approach with the standard flow cytometry method performed at the University of Washington. This was done using pre- and post-treatment paired samples from 98 B-ALL cases from the Children’s Oncology Group AALL0932 trial. Said Dr. Robins, "This allowed a direct comparison of our approach to the state of the art measurement of residual disease from one of the premier monitoring labs in the country." In 92 of the 98 pre-treatment samples, the authors were able to identify the unique, recombined IGH gene sequences that represented each patient’s clonal, neoplastic B lymphoblasts. The presence of these original clonal rearrangements could then be looked for in post-treatment samples to identify MRD.
Comparing the two methods, both detected MRD in 23 samples and did not detect MRD in 40 samples. However, there were 28 cases where MRD was detected by the IGH sequencing method but not flow cytometry (see figure). There were no cases where flow cytometry detected MRD but sequencing did not. Though a few of these detection-discordant samples were within the detection limit for flow cytometry, the authors estimated that their method had a 10-fold lower detection limit for MRD, at around 1 cell in a million. Since early detection of MRD is important for personalized treatment of B-ALL, this more sensitive detection could potentially improve patient prognostication. Said Robins, "These results are very encouraging, and suggest that our ClonoSeq technology can be translated effectively to the clinic."
Overall, this new method is a promising approach for the detection of minimal residual disease in B-ALL patients, particularly given its strong performance and ease of use compared to current methods. Additional studies will be needed, however, to understand the clinical implications of this more sensitive test. Said Robins, "Moving forward, we need to extend the study to much larger cohorts to evaluate differences in clinical outcome for low level residual disease (levels not detectable by presently employed methods). Additionally, we are in the process of applying the technology to many other types of leukemia and lymphoma. We hope to see a rapid uptake of ClonoSeq in the diagnostic market over the next couple years."
Illustration: Comparison of the clone frequencies identified by high-throughput sequencing (HTS, red) versus multi-parametric flow cytometry (mpFC, blue), for detection of minimal residual disease (MRD) in 29-day post-treatment B cell acute lymphoblastic leukemia cases (x-axis). The two methods both detected MRD for 23 samples (right), while for 28 samples MRD was detected using HTS but not mpFC (left). –Dr. Harlan Robins.
Read more…
Fred Hutchinson Cancer Research Center News Release (08/18/14)
Abstract (Clinical Cancer Research; 20(17), 4540-4548 (09/01/14))
