Genes Physically Held in Silencing 'Lock-Down' in Embryonic Stem Cells
The research presents how key decision-making genes which specify the embryo’s blueprint for subsequent development are physically clustered in the nucleus of embryonic stem cells and maintained in a silent state.
Researchers at the Babraham Institute have discovered a strong physical gene interaction network that is responsible for holding genes in a silencing grip during early development. In the same way that people can interact with others in close proximity, say within the same room, or others millions of miles apart, there are also short- and long-range interactions within the genome forming a three-dimensional configuration where different parts of the genome come into contact with each other. The research presents how key decision-making genes which specify the embryo’s blueprint for subsequent development are physically clustered in the nucleus of embryonic stem cells and maintained in a silent state.
The different cell types forming an embryo are derived from embryonic stem cells (ESCs). These cells are self-renewing and are maintained in an undifferentiated state meaning that they have the potential to become any cell type in the body. To become a specific cell type, embryonic stem cells progress along a developmental pathway, losing their stem cell characteristics and gaining new features. At a genomic level, assuming a specialized cellular identity reflects the switching on of appropriate developmental genes. Conversely, maintaining a stem cell identity requires the repression of developmental genes.
Using a novel technique developed at the Babraham Institute (Promoter Capture Hi-C), the researchers identified an unusually strong 3D network of developmental genes in ESCs. These genes encode proteins that establish the embryo’s body plan and direct organ development. As an ESC, you don’t want these instructions being read at this stage and so to prevent this, the genes are clustered together and silenced. The research showed that at the heart of this repression is a protein complex called Polycomb repressive complex (PRC1), a master regulator of ESC genome architecture. The research therefore establishes a mechanism, acting by physical interaction between specific genes and PRC1, which effectively holds genes in a silenced state. This prevents their expression in ESCs and so ensures maintenance of the undifferentiated state.
The researchers propose that the selective release of genes from this network leads to their expression and thus controls early development decisions that start the stem cell along the road to becoming a defined cell type. Lastly, de-regulation of Polycomb complexes has been shown to be the cause of several cancers and developmental disorders emphasizing the importance of understanding Polycomb-mediated gene repression in development and disease.
Dr. Sarah Elderkin, Group Leader in the Institute’s Nuclear Dynamics research program and lead author on the paper said: “Analyzing the genome-wide connections of 22,225 promoters in the genome of mouse embryonic stem cells allowed us to identify a sub-set of nearly 100 promoters which form the strongest interaction network seen in the entire genome. This is exciting because the members of this sub-set encode early developmental regulators which define what the embryonic stem cell will become. This research uncovers a mechanism for how inappropriate expression of developmental genes is prevented and also suggests how genes are freed from this silencing in order for normal embryonic development to proceed.”
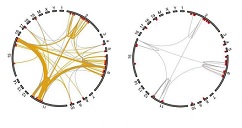
Illustration: Left-hand figure: the wild type mouse ESC genome; right-hand figure: reduced interactions in a mouse ESC genome from mouse ESCs lacking PRC1. The thickness of the lines relates to strength of interactions. Loss of the yellow lines demonstrates loss of biologically significant interactions. –Schoenfelder et al. Nature Genetics, 2015.
Read more…
Babraham Institute News Release (09/01/15)
AlphaGalileo (08/27/15)
Science Daily (08/31/15)
MedicalXpress (08/31/15)
Abstract (Nature Genetics; (08/31/15))
The different cell types forming an embryo are derived from embryonic stem cells (ESCs). These cells are self-renewing and are maintained in an undifferentiated state meaning that they have the potential to become any cell type in the body. To become a specific cell type, embryonic stem cells progress along a developmental pathway, losing their stem cell characteristics and gaining new features. At a genomic level, assuming a specialized cellular identity reflects the switching on of appropriate developmental genes. Conversely, maintaining a stem cell identity requires the repression of developmental genes.
Using a novel technique developed at the Babraham Institute (Promoter Capture Hi-C), the researchers identified an unusually strong 3D network of developmental genes in ESCs. These genes encode proteins that establish the embryo’s body plan and direct organ development. As an ESC, you don’t want these instructions being read at this stage and so to prevent this, the genes are clustered together and silenced. The research showed that at the heart of this repression is a protein complex called Polycomb repressive complex (PRC1), a master regulator of ESC genome architecture. The research therefore establishes a mechanism, acting by physical interaction between specific genes and PRC1, which effectively holds genes in a silenced state. This prevents their expression in ESCs and so ensures maintenance of the undifferentiated state.
The researchers propose that the selective release of genes from this network leads to their expression and thus controls early development decisions that start the stem cell along the road to becoming a defined cell type. Lastly, de-regulation of Polycomb complexes has been shown to be the cause of several cancers and developmental disorders emphasizing the importance of understanding Polycomb-mediated gene repression in development and disease.
Dr. Sarah Elderkin, Group Leader in the Institute’s Nuclear Dynamics research program and lead author on the paper said: “Analyzing the genome-wide connections of 22,225 promoters in the genome of mouse embryonic stem cells allowed us to identify a sub-set of nearly 100 promoters which form the strongest interaction network seen in the entire genome. This is exciting because the members of this sub-set encode early developmental regulators which define what the embryonic stem cell will become. This research uncovers a mechanism for how inappropriate expression of developmental genes is prevented and also suggests how genes are freed from this silencing in order for normal embryonic development to proceed.”
Illustration: Left-hand figure: the wild type mouse ESC genome; right-hand figure: reduced interactions in a mouse ESC genome from mouse ESCs lacking PRC1. The thickness of the lines relates to strength of interactions. Loss of the yellow lines demonstrates loss of biologically significant interactions. –Schoenfelder et al. Nature Genetics, 2015.
Read more…
Babraham Institute News Release (09/01/15)
AlphaGalileo (08/27/15)
Science Daily (08/31/15)
MedicalXpress (08/31/15)
Abstract (Nature Genetics; (08/31/15))
